Mastering High-Dimensional Data
At Cortexa Genomix, we have profound expertise in navigating the complexities of transcriptomics. RNA-Seq data, with its immense dimensionality often exceeding 19,000 features from just a handful of samples, presents a significant "curse of dimensionality" challenge. Our team expertly mitigates this by applying a suite of advanced dimensionality reduction techniques tailored to the unique structure of your biological data.
We leverage both unsupervised methods like Principal Component Analysis (PCA), Multi-Dimensional Scaling (MDS), and Autoencoders, alongside supervised methods such as ANOVA and Chi-Square analysis. This dual approach allows us to reduce complexity while preserving the critical biological signals necessary for building robust and accurate machine learning models. Our rigorous comparative analysis, as demonstrated in our work with Alzheimer's Disease data, ensures we select the optimal feature set for downstream classification.
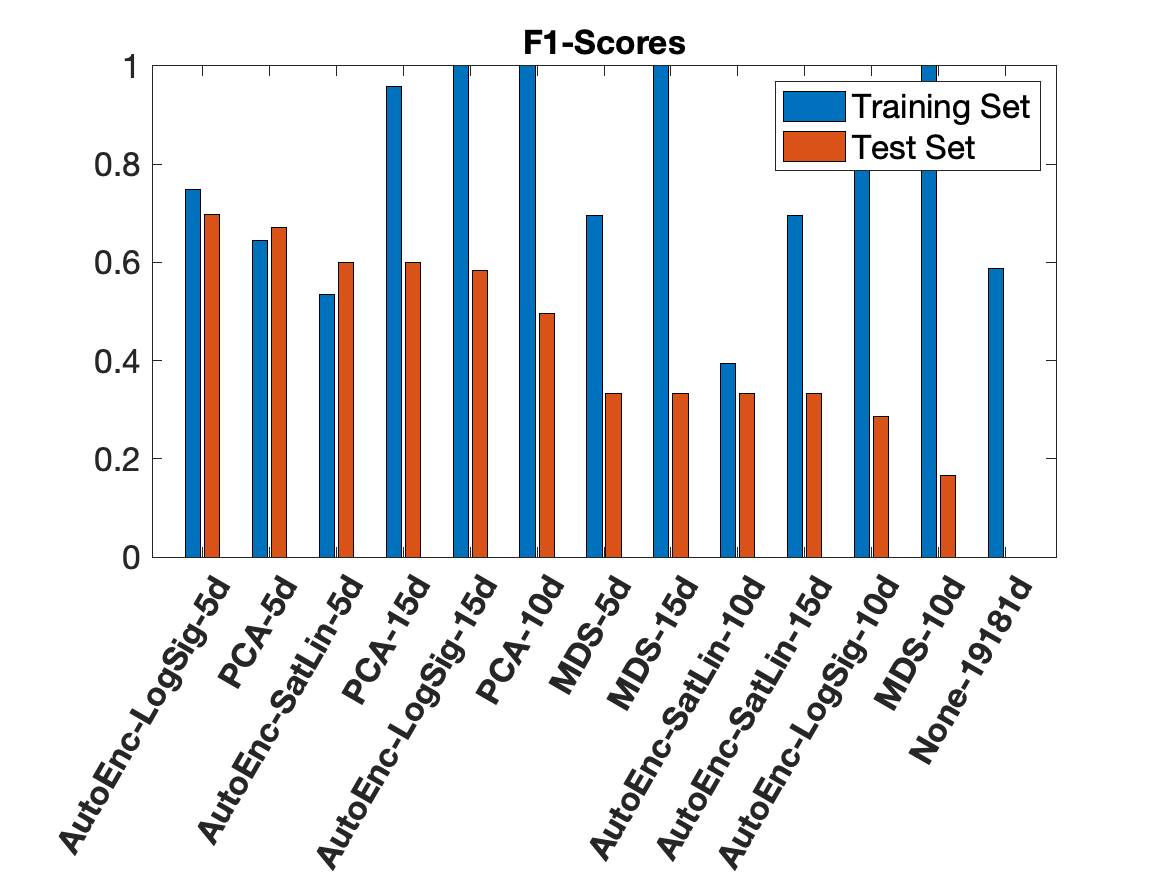
Comparative F1 Scores for dimensionality reduction on a 19,181-feature Alzheimer's RNA-Seq dataset